Projects
Ongoing Bioinformatics Projects
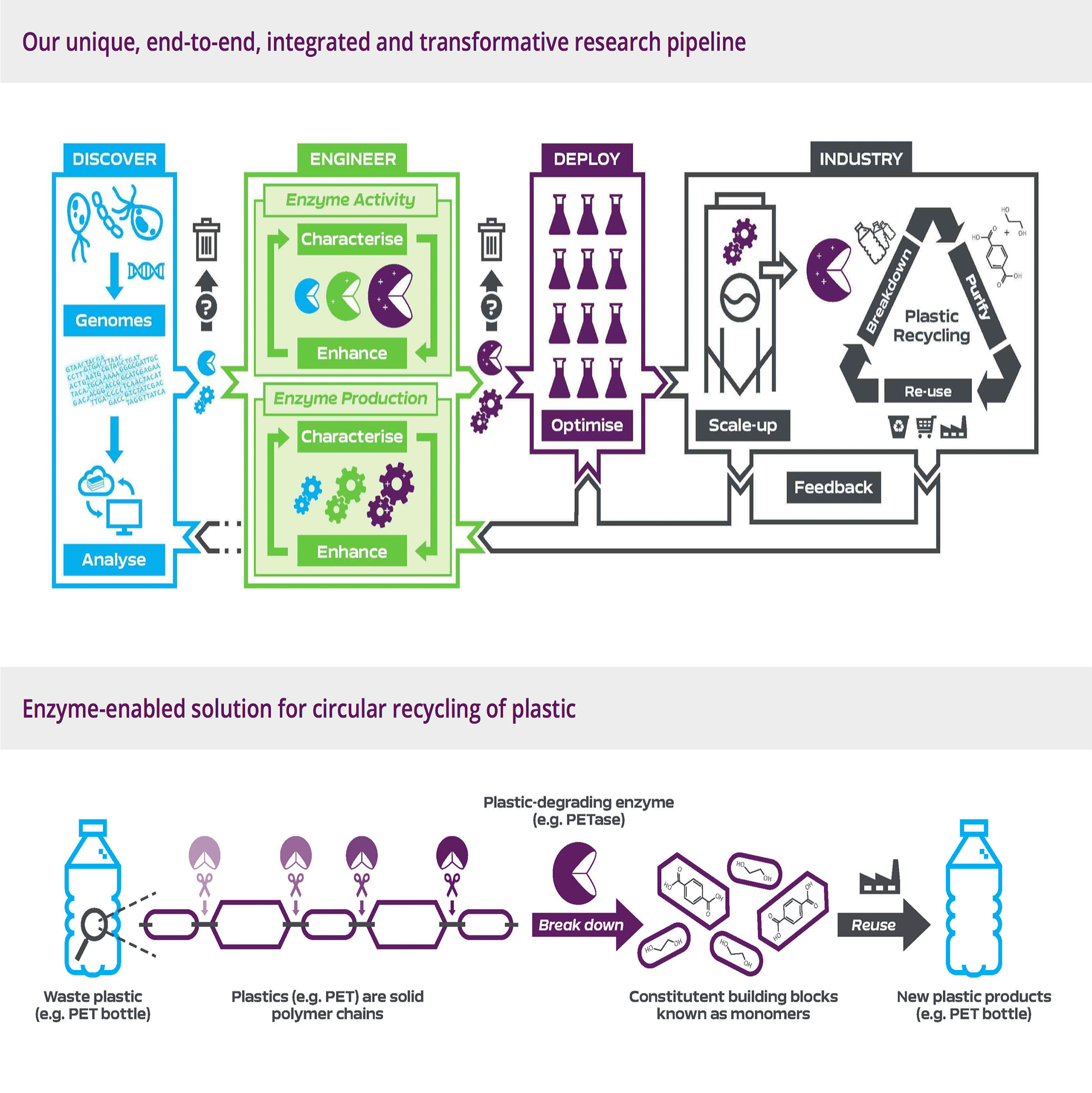
Center for Enzyme Innovation
An innovative new Research Centre to identify novel enzymatic solutions to environmental waste problems such as plastic

Portsmouth Heritage Hub
A network for researchers and stakeholders in local historical monuments and buildings to develop collaborative research projects with an aim to conserve, interpret, preserve and educate about our local heritage

Changes in microbial communities of Cliona celeta during observed changes in sponge health
Analysis of changes in microbial communities of Cliona celeta from Skomer Marine Nature Reserve during observed changes in sponge health using 16s rDNA sequencing

Genotyping of Ancient DNA from Crew Members from the Mary Rose
Genotyping of ancient DNA from crew members from the Mary Rose to identfy phenotypic traits and disease traits

Development of Bioinformatics Tools
We are currently working on a number of in-house bioinformatics tools to make these available to the wider research community

Effects of Anti-Fouling Coatings on Marine Biofilms
Metatranscriptomic analysis of marine biofilm composition on commercially available and novel anti-fouling substrates

Effects of Radiation Exposure in the Environment
Identification of differentially regulated gene pathways as a result of radiation exposure
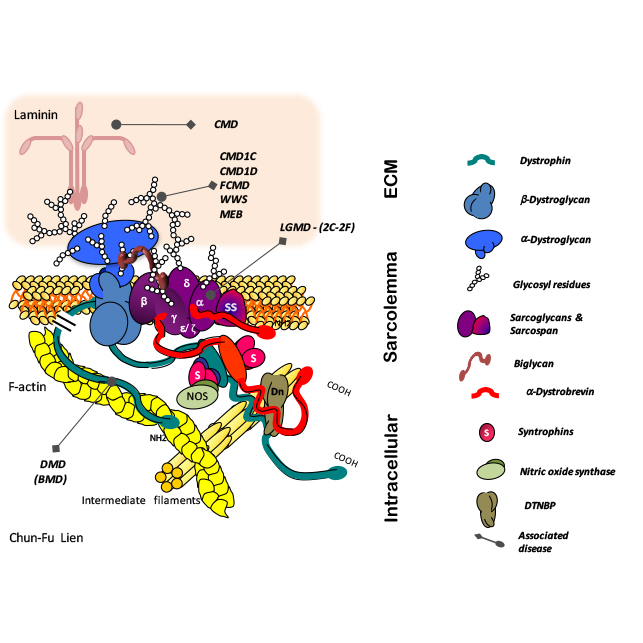
Gene Expression Profiling of Duchenne Muscular Dystrophy
Differential gene expression analysis in a model of Duchenne Muscular Dystrophy

The Role of Variant Histones in Xenopus Development
Transcriptional profiling of variant histone knockdowns in Xenopus laevis

Transcriptome Profiling Of The Radula of Patella vulgata
Identification of key gene pathways involved in the formation of the radula in Patella vulgata (common limpet)